How to evaluate an assembly
VirAmp uses QUAST (Quality Assessment Tools for Genome Assemblies) to evaluate the newly assembled genome.
Basic statistics such as contig lengths, N50, NG50, # misassemblies etc. are given on screen, and more detailed report is provided for download on the right "history" panel.
Two important statistics here are N50 and NG50.
- N50 is a weighted median statistic such that 50% of the entire assembly is contained in contigs or scaffolds equal to or larger than this value (Rounsley, S. etc. 2009). This is one of the most accepted statistics to evaluate the quality of assembly.
- NG50 is a modified version of N50. When a reference genome is presented, the length of reference genome is used for normalization instead of the cumulative length of the entire assembly. This reduces the bias brought by misassemlies, which may cause trouble when the percentage of misassemblies is significant.
What to input
- Reference genome in FASTA format
- Target genome: Your newly assembled genome (or other genome you want to compare with the referece) in FASTA format
- Contig length threshold: Minimum length of contigs you want to consider as valid, by default is 500 nucleotides.
What is shown on screen:
Assembly viramp # contigs (>= 0 bp) 5 # contigs (>= 1000 bp) 5 Total length (>= 0 bp) 135722 Total length (>= 1000 bp) 135722 # contigs 5 Largest contig 108094 Total length 135722 Reference length 136376 GC (%) 67.36 Reference GC (%) 67.53 N50 108094 NG50 108094 N75 108094 NG75 108094 # misassemblies 0 # local misassemblies 3 # unaligned contigs 0 + 1 part Unaligned contigs length 95 Genome fraction (%) 98.610 Duplication ratio 1.009 # N's per 100 kbp 106.84 # mismatches per 100 kbp 472.19 # indels per 100 kbp 72.87 Largest alignment 107807 NA50 107807 NGA50 107807 NA75 107807 NGA75 107807
Featured graphs in the download version
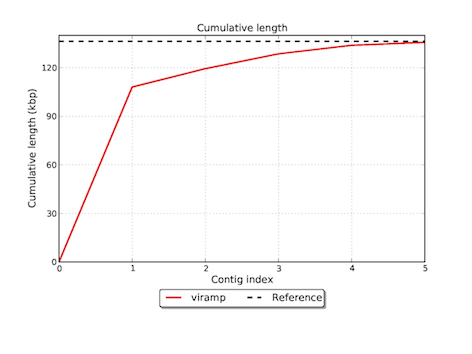
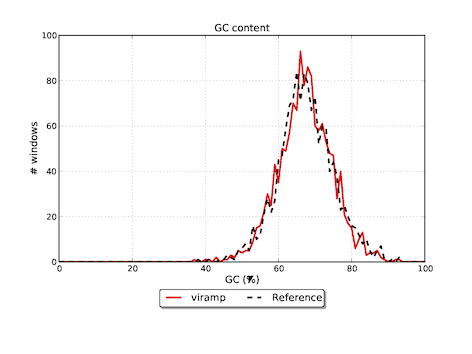
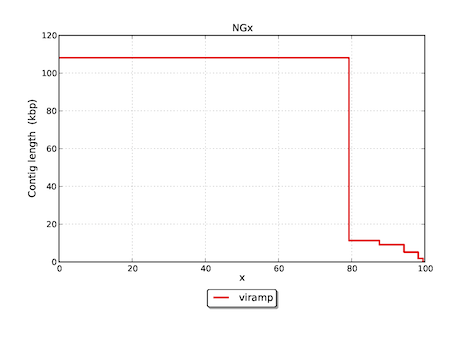
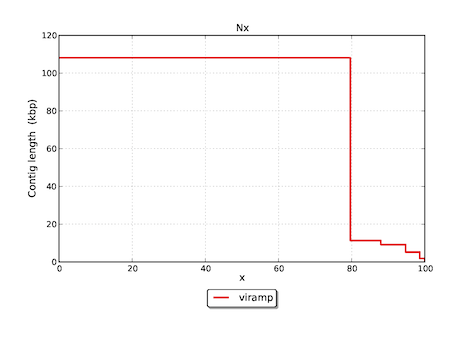
Reference
QUAST: quality assessment tool for genome assemblies. Alexey Gurevich, Vladislav Saveliev, Nikolay Vyahhi and Glenn Tesler. Bioinformatics (2013)29(8):1072-1075.